Case 1 Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns
Abstract
While splicing changes caused by somatic mutations in SF3B1 are known, identifying fulllength isoform changes may better elucidate the functional consequences of these mutations.
We report nanopore sequencing of full-length cDNA from CLL samples with and withoutSF3B1 mutation, as well as normal B cell samples, giving a total of 149 million pass reads. We present FLAIR (Full-Length Alternative Isoform analysis of RNA), a computational workflow to identify high-confidence transcripts, perform differential splicing event analysis, and differential isoform analysis. Using nanopore reads, we demonstrate differential 3’ splice site changes associated with SF3B1 mutation, agreeing with previous studies. We also observe a strong downregulation of intron retention events associated with SF3B1 mutation. Full-length transcript analysis links multiple alternative splicing events together and allows for better estimates of the abundance of productive versus unproductive isoforms. Our work demonstrates the potential utility of nanopore sequencing for cancer and splicing research.
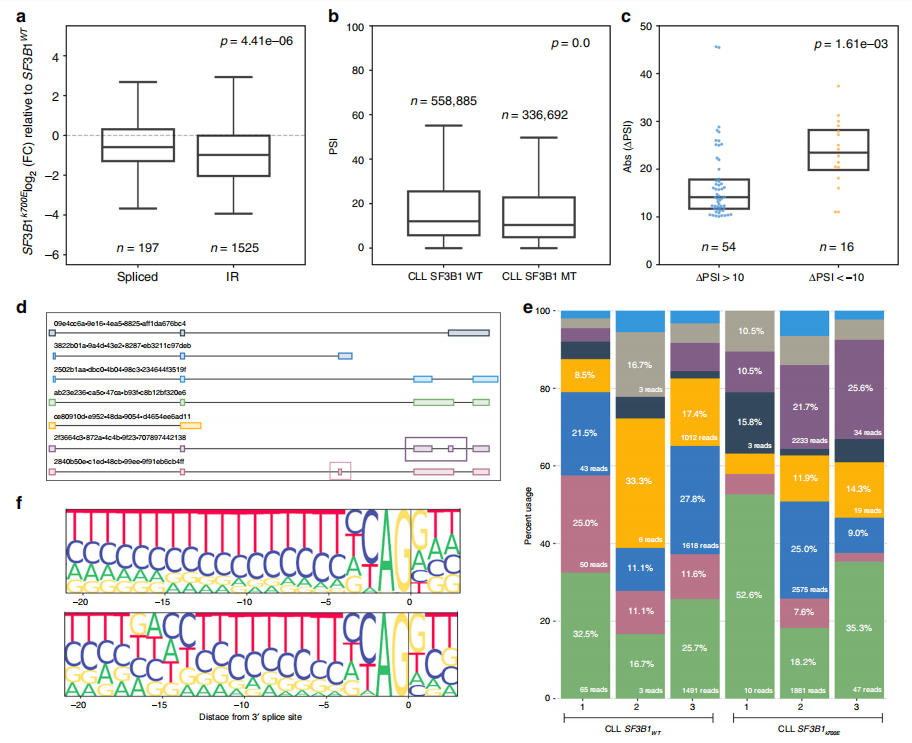
Fig 1 Intron retention events are more strongly downregulated in CLL SF3B1K700E
References
Tang AD. et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nature Communications,2020
Case 2 Nanopore sequencing reveals full-length Tropomyosin 1 isoforms and their regulation by RNA-binding proteins during rat heart development
Abstract
Alternative splicing (AS) contributes to the diversity of the proteome by producing multiple isoforms from a single gene. Although short-read RNA-sequencing methods have been the gold standard for determining AS patterns of genes, they have a difficulty in defining full-length mRNA isoforms assembled using different exon combinations. Tropomyosin 1 (TPM1) is an actin-binding protein required for cytoskeletal functions in non-muscle cells and for contraction in muscle cells. Tpm1 undergoes AS
regulation to generate muscle versus non-muscle TPM1 protein isoforms with distinct physiological functions. It is unclear which full-length Tpm1 isoforms are produced via AS and how they are regulated during heart development. To address these, we utilized nanopore long-read cDNA sequencing without gene-specific PCR amplification. In rat hearts, we identified full-length Tpm1 isoforms composed of distinct exons with specific exon linkages. We showed that Tpm1 undergoes AS transitions during embryonic heart development such that muscle-specific exons are connected generating predominantly muscle-specific Tpm1 isoforms in adult hearts. We found that the RNA-binding protein RBFOX2 controls AS of rat Tpm1 exon 6a, which is important for cooperative actin binding. Furthermore, RBFOX2 regulates Tpm1 AS of exon 6a antagonistically to the RNA-binding protein PTBP1. In sum, we defined full-length Tpm1 isoforms with different exon combinations that are tightly regulated during cardiac development and provided insights into the regulation of Tpm1 AS by RNA-binding proteins. Our results demonstrate that nanopore sequencing is an excellent tool to determine full-length AS variants of muscle-enriched genes.
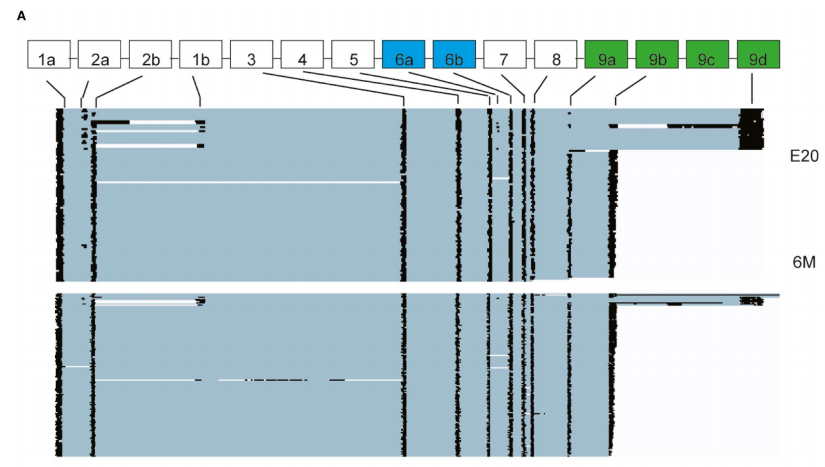
Fig 1 Identification of full-length Tpm1 isoforms generated via alternative splicing during rat heart development using nanopore
sequencing
References
Jun Cao. et al. Nanopore sequencing reveals full-length Tropomyosin 1 isoforms and their regulation by RNA binding proteins during rat heart development. BioRxiv,2020