Classic case:
Title:
A genomic variation map provides insights into the genetic basis of spring chinese cabbage (Brassica rapa ssp. pekinensis) selection
Journal:
Molecular Plant (IF: 13.164)
Background:
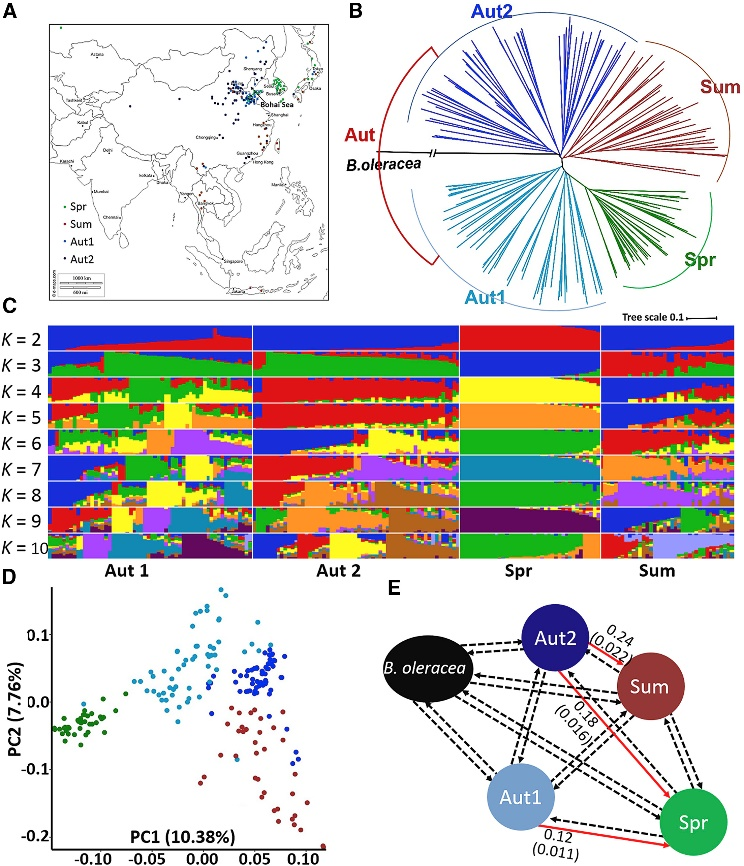
The domestication of Chinese cabbage can be traced back to about 6000 years ago. Based on genetic and taxonomic analyses, autumn cabbage originated in the temperate regions of central China and was subsequently introduced to northern and southern China as well as to Korea and Japan. In southern China, a heat-tolerant summer cabbage variety was selected to adapt to the subtropical environment. About 40 years ago, a number of late spring cabbage varieties were bred in Korea and Japan and introduced back to China. These varieties are well adapted to the cold climate of early spring in China. However the following questions are unclear: (1) which type of cabbage is the ancestor of spring cabbage; (2) where is the geographical origin of spring cabbage; and (3) what is the genetic basis of spring cabbage selection? In this study, we performed genetic evolutionary analysis based on resequencing to elucidate the basis of genetic variation in spring cabbage during domestication.
Key finding:
Chinese cabbage is the most consumed leafy crop in East Asian countries. However, premature bolting induced by continuous low temperatures severely decreases the yield and quality of the Chinese cabbage, and therefore restricts its planting season and geographic distribution. In the past 40 years, spring Chinese cabbage with strong winterness has been selected to meet the market demand. Here, we report a genome variation map of Chinese cabbage generated from the resequencing data of 194 geographically diverse accessions of three ecotypes. In-depth analyses of the selection sweeps and genome-wide patterns revealed that spring Chinese cabbage was selected from a specific population of autumn Chinese cabbage around the area of Shandong peninsula in northern China. We identified 23 genomic loci that underwent intensive selection, and further demonstrated by gene expression and haplotype analyses that the incorporation of elite alleles of VERNALISATION INSENTIVE 3.1 (BrVIN3.1) and FLOWER LOCUS C 1 (BrFLC1) is a determinant genetic source of variation during selection. Moreover, we showed that the quantitative response of BrVIN3.1 to cold due to the sequence variations in the cis elements of the BrVIN3.1 promoter significantly contributes to bolting-time variation in Chinese cabbage. Collectively, our study provides valuable insights into the genetic basis of spring Chinese cabbage selection and will facilitate the breeding of bolting-resistant varieties by molecular-marker-assisted selection, transgenic or gene editing approaches.
Publication
Su TB, Wang WH, Li PR et al. A genomic variation map provides insights into the genetic basis of spring chinese cabbage (Brassica rapa ssp. pekinensis) selection. Molecular Plant, 2018.