- Product introduction
- Common problem
- Classic case
- Result display
- Circular RNA (circRNA) is a new class of non-coding RNAs that are widely present in plants and animals. By enriching circRNA by removing rRNA and linear RNA by strand-specific library building method, based on high-throughput sequencing, a large amount of circRNA information can be obtained, and the structure and target target of cirRNA can be predicted through bioinformatics analysis
Product features
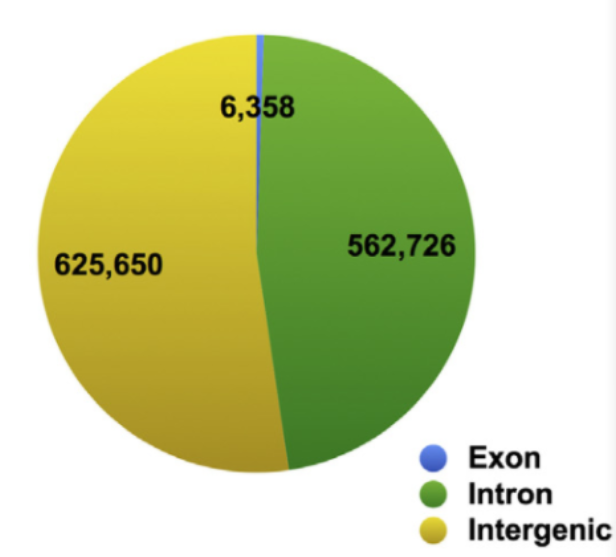
Identify circRNA transcripts by circRNA cyclic splicing and analyze the source of circRNA.
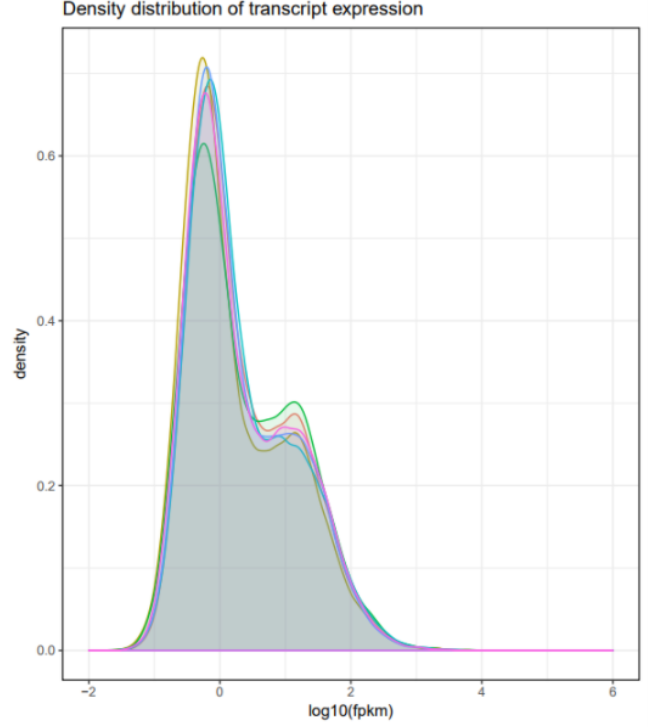
The expression of circRNA is generally tissue specific, and by estimating the amount of circRNA expression in all samples, it is possible to understand the expression pattern of circRNA in the sample, and the correlation between fractal samples in the expression pattern.
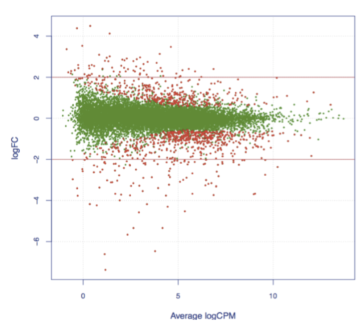
Between the treatment group and the control group, the differentially expressed circRNA was analyzed, and the relationship between the differential circRNA and the phenotypic difference was speculated.
KEGG/GO enrichment of source genes
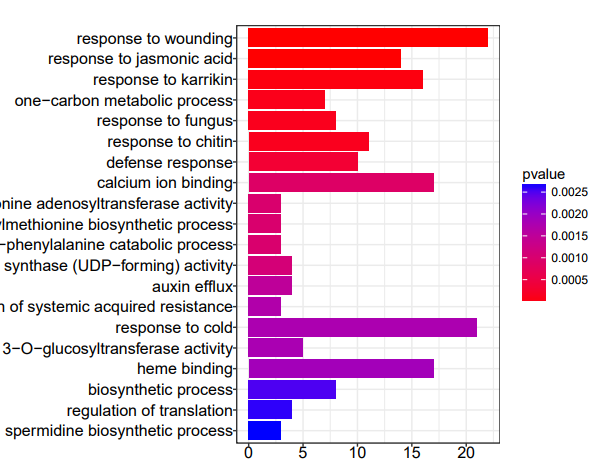
Transcripts of genes that exist under certain conditions form circRNA instead of mRNA. GO/KEGG enrichment analysis of the source genes of differential circRNAs is helpful in analyzing the differential phenotype and circRNA function.
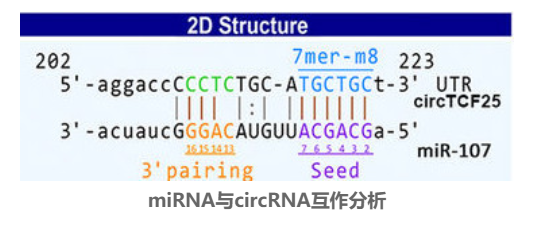
Studies have found the presence of miRNAs binding locations on some circRNA sequences, which can recruit miRNAs and participate in transcriptional regulation.
-
Q1.What are the characteristics of the circRNA library building method?
A:The circRNA library method uses the strategy of removing rRNA and removing linear RNA to enrich circRNA, and uses strand specific library to distinguish between the positive and negative strands of circRNA sources and genes.
Q2.How much data should each sample be measured for repetition?
A:Recommend the amount of data greater than 8G. If comparative analysis between samples is required, at least 3 intra-group replicates are recommended.
-
Case Widespread noncoding circular RNAs in plants
Abstract
A large number of noncoding circular RNAs (circRNAs) with regulatory potency have been identified in animals, but little attention has been given to plant circRNAs.
We performed genome-wide identification of circRNAs in Oryza sativa and Arabidopsis thaliana using publically available RNA-Seq data, analyzed and compared features of plant and animal circRNAs.
circRNAs (12037 and 6012) were identified in Oryza sativa and Arabidopsis thaliana, respectively, with 56% (10/18) of the sampled rice exonic circRNAs validated experimentally. Parent genes of over 700 exonic circRNAs were orthologues between rice and Arabidopsis, suggesting conservation of circRNAs in plants. The introns flanking plant circRNAs were much longer than introns from linear genes, and possessed less repetitive elements and reverse complementary sequences than the flanking introns of animal circRNAs. Plant circRNAs showed diverse expression patterns, and 27 rice exonic circRNAs were found to be differentially expressed under phosphate-sufficient and -starvation conditions. A significantly positive correlation was observed for the expression profiles of some circRNAs and their parent genes.
Our results demonstrated that circRNAs are widespread in plants, revealed the common and distinct features of circRNAs between plants and animals, and suggested that circRNAs could be a critical class of noncoding regulators in plants.
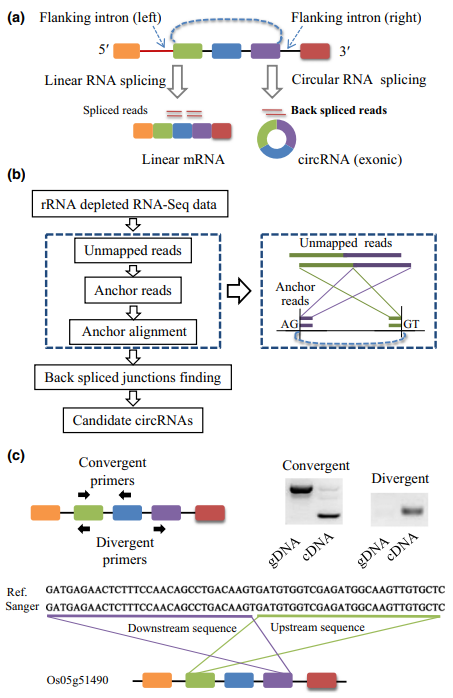
Fig 1 Identification of circular RNAs (circRNAs) in plants
References
Chu Yu Ye, et al. Widespread noncoding circular RNAs in plants. New Phytologist,2015
-
Copyright © 2018 Wuhan Benagen Technology Co., Ltd . All Rights Reserved.




