Successful Case
Study on the mechanisms of the cross-resistance to TET, PIP and GEN in Staphylococcus aureus mediated by the Rhizoma Coptidis extracts
Abstract
The purpose of this study was focused on the mechanisms of the cross-resistance to tetracycline (TET), piperacillin Sodium (PIP), and gentamicin (GEN) in Staphylococcus aureus (SA) mediated by Rhizoma Coptidis extracts (RCE). The selected strains were exposed continuously to RCE at the sublethal concentrations for 12 days, respectively. The susceptibility change of the drug-exposed strains was determined by analysis of the minimum inhibitory concentration. The 16S rDNA sequencing method was used to identify the RCE-exposed strain. Then the expression of resistant genes in the selected isolates was analyzed by transcriptome sequencing. The results indicated that RCE could trigger the preferential crossresistance to TET, PIP, and GEN in SA. The correlative resistant genes to the three kinds of antibiotics were upregulated in the RCE-exposed strain, and the mRNA levels of the resistant genes determined by RT-qPCR were consistent with those from the transcriptome analysis. It was suggested from these results that the antibacterial Traditional Chinese Medicines might be a significant factor of causing the bacterial antibiotic-resistance.
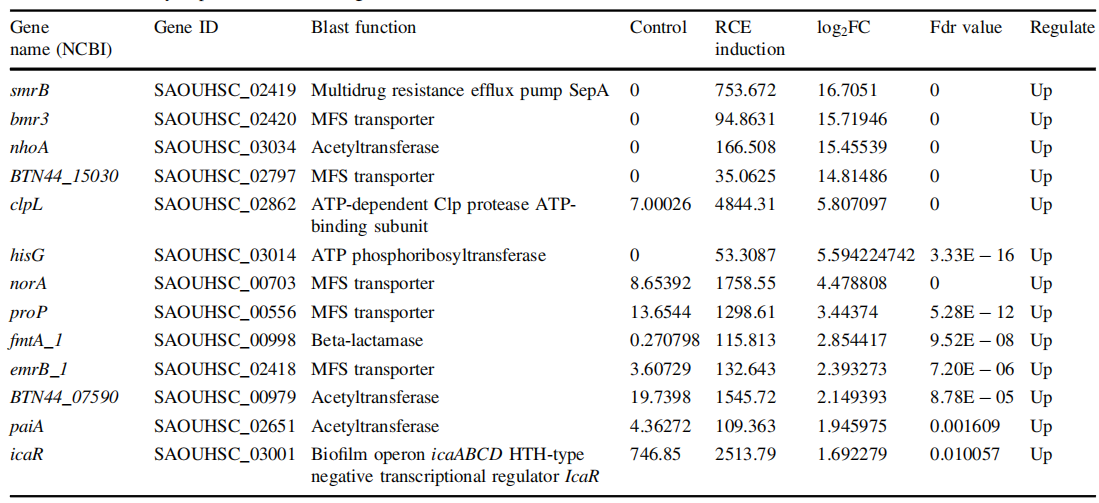
Table Differentially expression of resistant genes
 References
References
Sugui Lan, et al. Study on the mechanisms of the cross-resistance to TET, PIP and GEN in Staphylococcus aureus mediated by the Rhizoma Coptidis extracts.The Journal of Antibiotics,2021