- Product introduction
- Common problem
- Classic case
- Result display
- Product introduction
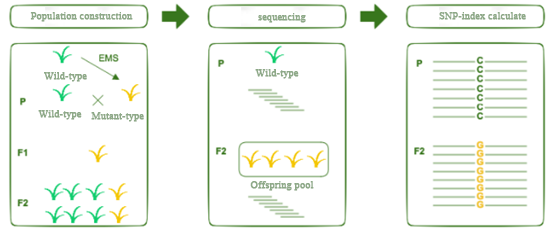
The wild-type plants were mutated by EMS to obtain the mutant type, and the mutant plants were crossed with the wild-type after multiple generations of self-fertilization to obtain F1, and F1 was self-fertilized to obtain F2, and F2 showed segregation of traits.
Several strains of F2 were selected for sequencing in a mixed pool of individuals with phenotypic inconsistency with the wild type, while the wild type sequencing was used as the reference gene sequence.
SNP-Index (the proportion of reads different from the reference sequence to the total reads) significantly higher than the desired SNP site was used as a candidate site to cause the mutation.
-
FAQ:
(1) Is DNA extracted from each individual separately and then mixed pool or is the sample mixed pool and then DNA extracted?
A: Extracting DNA first and then mixing in equal amounts reduces systematic errors. Some literature mentions that there is no significant difference between the two methods, but no data support is given. It is recommended to choose with the actual situation.
(2) What if the mutant cannot be crossed with the wild type?
A: In addition to the standard procedure of MutMap, it can be flexibly adapted in practical applications, as long as sufficient recessive pure offspring can be generated. For example, when the mutant cannot be crossed with the wild type, the mutant strain can be self-crossed to obtain a pool of offspring.
(3) How many individuals should be included in the offspring pool? What is the sequencing depth?
A: Number of individuals: 20 to 50, 30 recommended; Sequencing depth: 20X to 50X, 30X recommended.
(4) How to reduce false positives?
A: During EMS mutagenesis, in addition to causing Causal mutation, other SNP variants are also generated. Some of these variants will be purified and retained during self-crossing and cannot be distinguished from Causal mutation in the analysis.R Fekih proposed MutMap+ in 2013, which enables mutants to segregate traits by self-crossing. Using the segregated individuals, separate pools were mixed and sequenced. Noise SNPs were eliminated by comparing SNP-Index differences.
-
Classic case:
Title:
MutMap accelerates breeding of a salt-tolerant rice cultivar.
Journal:
Nature Biotechnology (54.908)
Background:
The 2011 earthquake and tsunami caused salt contamination in more than 20,000 hectares of rice fields in Japan caused by seawater inundation. Due to the lack of local rice varieties tolerant to high salt concentration in Japan, there is an urgent need to breed new salt-tolerant rice varieties. The authors obtained the salt-tolerant mutant hst1 by EMS mutagenesis of the local variety Hitomebore. By MutMap method, the SNPs leading to gene inactivation were localized, which led to salt tolerance. A new salt-tolerant variety Kaijin was produced, which differs from Hitomebore by only 201 SNPs. Its growth and yield were consistent with Hitomebore. The entire breeding process of the new variety took only 2 years.
Key findings:
(1) the SNP-Index deviated significantly from 0.5 in the region 2.76-8.57M on chromosome 6,which contains two SNPs, one of which is located on the third exon of the gene OsRR22, which encodes a B-type response regulator. this SNP causes premature termination of the gene and loss of function.
(2) Demonstration by transposon insertion, and PCR experiments that inactivation of OsRR22can produce a salt-tolerant phenotype.
(3) Investigating gene expression differences by RNA-Seq technology and finding that themutation only affects the expression of a small number of genes, but does not clarify the mechanism leading to salt tolerance.
(4) The salt-tolerant mutant hst1 was used as the parent and Hitomebore was used as the father to
produce the salt-tolerant variety "Kaiji". This variety retained the excellent yield and quality traits of its parents while maintaining the same level of salt tolerance as the mutant hst1. The variety was promoted locally in 2015.
Publication
Hiroki T, Muluneh T, Akira A et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nature biotechnology, 2015
-
Copyright © 2018 Wuhan Benagen Technology Co., Ltd . All Rights Reserved.