Case study: joint analysis of multiomics reveals the regulatory effect of autophagy on Maize metabolism (if = 13.256)
Under nutritional constraints, autophagy is essential for cell life activities. ATG is a series of autophagy related proteins. Atg8 is a ubiquitin protein. Under the action of atg12-atg5-atg16, it binds with PE to form atg8-pe complex, which plays an important role in the process of autophagy.
scientific problems
Mechanism of cell dependent autophagy in Maize
experimental design
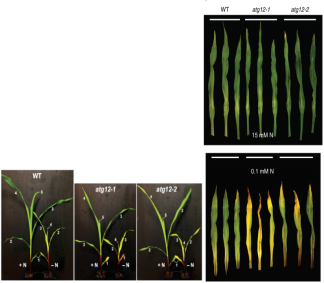
Wild type (W22), atg12-1 mutant (Atg8 modification was blocked and inactivated), and atg12-2 (gene knockout) were cultured under nitrogen rich conditions for 2 weeks and under nitrogen deficient conditions for 1 week, respectively. The second and fourth leaves of seedlings at two stages were taken for transcriptome (PCR verification), proteome, metabolome and Ionome detection.
result
Phenotypic nitrogen stress changed the transcriptome and proteome of WT leaves, increased nitrogen metabolism and decreased chlorophyll expression. Nitrogen deficient seedlings showed strong growth inhibition and atg12 mutant senescence.
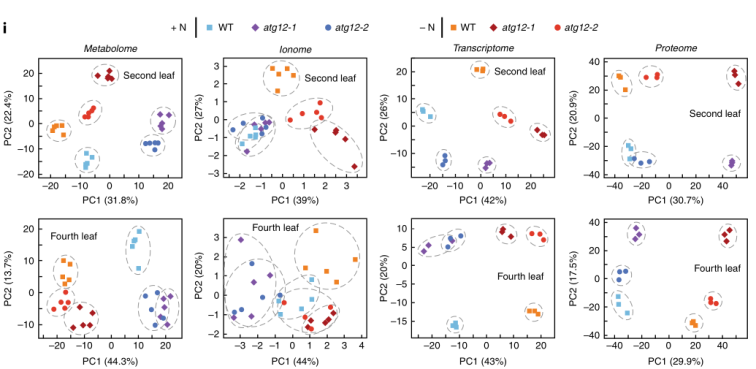
There were differences in transcriptome, proteome and metabolome (PCA) between multiomic mutants and wild-type under nitrogen rich or nitrogen deficient conditions.
The abundance of proteome Atg8 protein family and total protein increased in atg12, indicating that autophagy is important to maintain normal protein abundance in plants. Atg12 mutant blocks the conversion mediated by Atg8 autophagy.
Transcriptome autophagy significantly affected maize transcriptome, and protease and lipid metabolism transcripts in autophagy mutants were significantly up-regulated in 567 core transcripts.
The metabolites of metabolome were significantly different under nitrogen rich conditions, indicating that autophagy could significantly affect maize metabolism even under normal growth conditions; The relatively small differences during stress, especially lipids, indicate that hunger activates other metabolic pathways other than autophagy to help meet this challenge.
association analysis
There was no significant change in the mRNA corresponding to most of the abundant proteins in atg12-1. On the surface, these changes were caused by autophagy and had nothing to do with synthesis. The increased levels of protein and metabolites are consistent with the increased levels of these metabolites. The consumption of thylakoids in mutants is consistent with the accelerated decomposition of chloroplasts, which may be due to the lack of other decomposition pathways under autophagy conditions.
The changes of major mRNA and protein and metabolites related to lipid metabolism in atg12 indicate that the excessive accumulation of lipid metabolites in the mutant is caused by defects in other metabolic processes regulated by autophagy, which may be related to protein clearance.
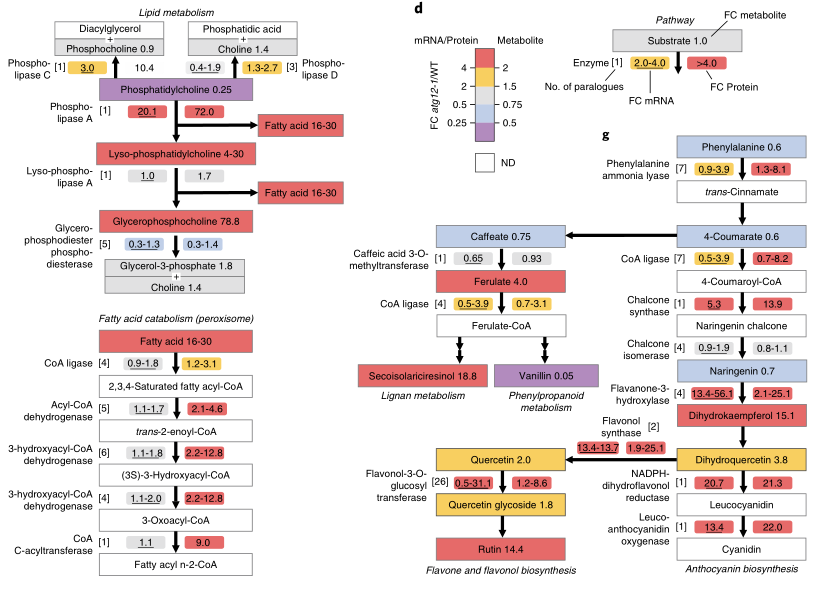
Conclusion autophagy essentially changes the protein composition and metabolites of maize, and provides nutrients for plant growth. It is an important part of maintaining cell homeostasis.
reference
Fionn M, Robert C A, Richard S M, et al. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nature Plants 4, 1056–1070 (2018).