- Product introduction
- Common problem
- Classic case
- Result display
- Full spectrum of space-time dynamic transcripts aims to construct an Isoform database of different genes of a species, analyze the Transcripts and variable splicing forms of different tissues, different parts, some special cells, and important periods, draw a fine map of the full-length transcriptome of cells /tissues /organs with spatio -temporal characteristics, reconstruct the dynamic network in the living system, and analyze the life process and activity law of a species from the transcript level.
Product advantages
v 1. Period/tissue-specific Transcripts identification
v 2. Dynamic function network construction based on the full spectrum of Transcripts
v 3. Establish a variable splicing database of species
Product application
Ø Developmental biology research and genetics transcriptional regulatory mechanism
Ø Disease transcriptional regulation mechanism study
Ø Important economic species Transcripts and variable splicing database construction
Ø Schema species reference Transcripts and variable splicing database build libraries
-
-
Transcriptome variation in human tissues revealed by long-read sequencing
Abstract
Regulation of transcript structure generates transcript diversity and plays an important role in human disease. The advent of long-read sequencing technologies offers the opportunity to study the role of genetic variation in transcript structure. In this paper, we present a large human long-read RNA-seq dataset using the Oxford Nanopore Technologies platform from 88 samples from GTEx tissues and cell lines, complementing the GTEx resource. We identified just under 100,000 new transcripts for annotated genes, and validated the protein expression of a similar proportion of novel and annotated transcripts. We developed a new computational package, LORALS, to analyze genetic effects of rare and common variants on the transcriptome via allele-specific analysis of long reads. We called allele-specific expression and transcript structure events, providing novel insights into the specific transcript alterations caused by common and rare genetic variants and highlighting the resolution gained from long-read data. We were able to perturb transcript structure upon knockdown of PTBP1, an RNA binding protein that mediates splicing, thereby finding genetic regulatory effects that are modified by the cellular environment. Finally, we use this dataset to enhance variant interpretation and study rare variants leading to aberrant splicing patterns.
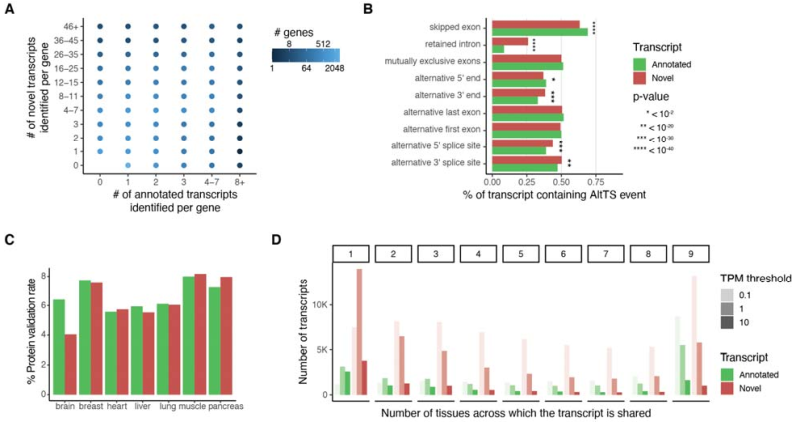
Fig 1 Discovery of new transcripts and comparison between tissues
References
Dafni A Glinos, et al. Transcriptome variation in human tissues revealed by long-read sequencing. BioRxiv,2021
-
Copyright © 2018 Wuhan Benagen Technology Co., Ltd . All Rights Reserved.