- Product introduction
- Common problem
- Classic case
- Result display
- Single cell RNA-seq
10x Genomics single-cell transcriptome sequencing (single cell RNA-seq, scRNA-seq) uses 10X Genomics combined with next-generation sequencing technology to label and distinguish a large number of cells that form tissues or organs, and perform high-level mRNA analysis at the single cell level. It can study the gene expression profile of a single cell with high-throughput single-molecule resolution, so as to reveal the heterogeneity of complex cell populations, At the same time, avoid that the gene expression signal of a single cell is masked by the average signal of the population.
Product advantages
True single cell level sequencing, not tissue level sequencing
Solve the problem of cell heterogeneity (analyze the expression characteristics of each cell)
Excavate important cell types and explore disease pathogenesis and related markers
Experimental principle
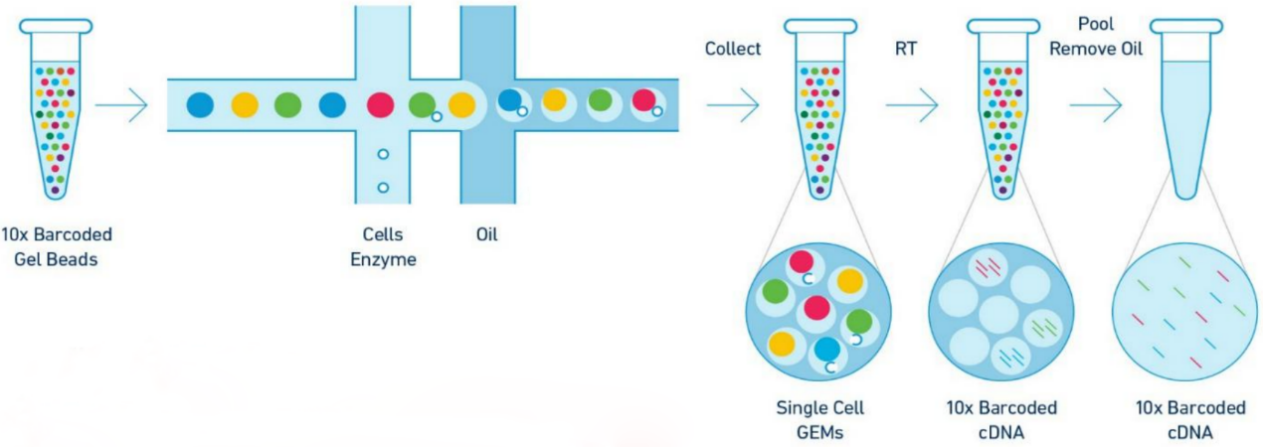
Application direction
Organ/tissue cell atlas
Developmental Biology Research
Disease-associated cell type Research
Tumor research
Virological research
-
-
Identification of a distinct luminal subgroup diagnosing and stratifying early stage prostate cancer by tissue-based single-cell RNA sequencing
Background: The highly intra-tumoral heterogeneity and complex cell origination of prostate cancer greatly limits the utility of traditional bulk RNA sequencing in finding better biomarker for disease diagnosis and stratification. Tissue specimens based single-cell RNA sequencing holds great promise for identification of novel biomarkers. However, this technique has yet been used in the study of prostate cancer heterogeneity.
Methods: Cell types and the corresponding marker genes were identified by single-cell RNA sequencing. Malignant states of different clusters were evaluated by copy number variation analysis and differentially expressed genes of pseudo-bulks sequencing. Diagnosis and stratification of prostate cancer was estimated by receiver operating characteristic curves of marker genes. Expression characteristics of marker genes were verified by immunostaining.
Results: Fifteen cell groups including three luminal clusters with different expression profiles were identified in prostate cancer tissues. The luminal cluster with the highest copy number variation level and marker genes enriched in prostate cancer-related metabolic processes was considered the malignant cluster. This cluster contained a distinct subgroup with high expression level of prostate cancer biomarkers and a strong distinguishing ability of normal and cancerous prostates across different pathology grading. In addition, we identified another marker gene, Hepsin (HPN), with a 0.930 area under the curve score distinguishing normal tissue from prostate cancer lesion. This finding was further validated by immunostaining of HPN in prostate cancer tissue array.

Fig. Diverse cell types in PCa were identified by single-cell sequencing.
REFERENCES
Ma X, et al. Identification of a distinct luminal subgroup diagnosing and stratifying early stage prostate cancer by tissue-based single-cell RNA sequencing. Mol Cancer, 2020;19(1):147.
-
Copyright © 2018 Wuhan Benagen Technology Co., Ltd . All Rights Reserved.
