Classic case:
Title:
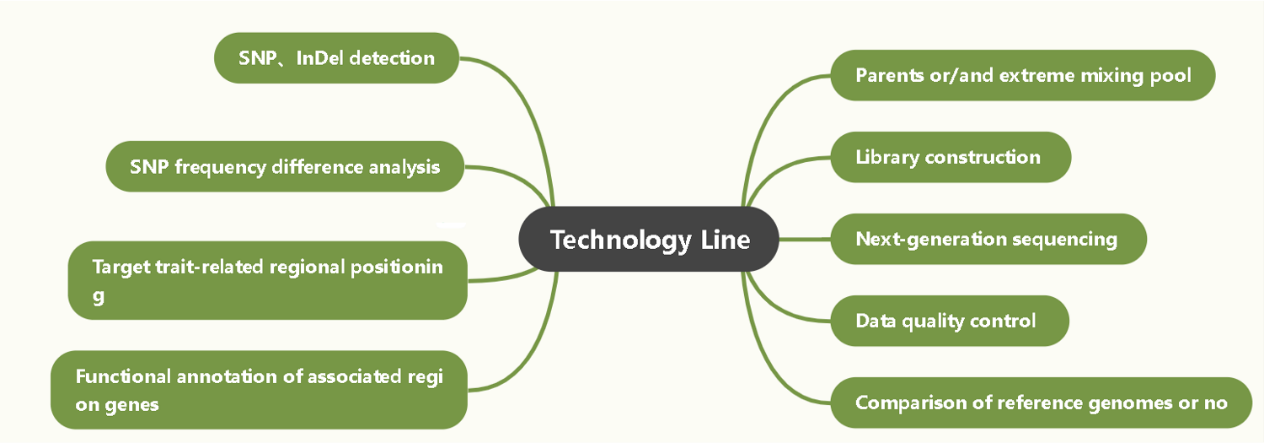
QTG-Seq Accelerates QTL Fine Mapping through QTL Partitioning and Whole-Genome Sequencing of Bulked Segregant Samples.
Journal:
Molecular Plant (IF: 13.164)
Background:
Agronomic traits are the direct goal of crop improvement breeding, a process that is critical for global food security. Modern crop improvement relies on the dissection of genetic and molecular mechanisms of agronomic traits. Figure cloning is an effective method for isolating QTGs, however it is time consuming and limited in its ability to detect rare alleles. Quantitative trait locus sequencing (QTLseq) has too low resolution to identify candidate genes, especially in species with large genome sizes. Therefore, there is still a need for a fine-grained strategy to rapidly identify candidate genes for potential quantitative traits. Maize is a widely studied genetic model, one of the most widely grown crops in the world, and its plant height traits can be used as a model for traits validated by QTG-seq methods.
Key finding:
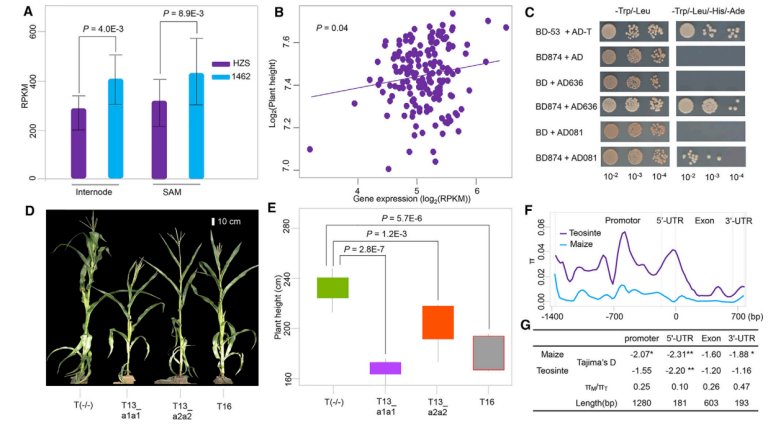
Deciphering the genetic mechanisms underlying agronomic traits is of great importance for crop improvement. Most of these traits are controlled by multiple quantitative trait loci (QTLs), and identifying the underlying genes by conventional QTL fine-mapping is time-consuming and labor-intensive. Here, we devised a new method, named quantitative trait gene sequencing (QTG-seq), to accelerate QTL fine-mapping. QTG-seq combines QTL partitioning to convert a quantitative trait into a near-qualitative trait, sequencing of bulked segregant pools from a large segregating population, and the use of a robust new algorithm for identifying candidate genes. Using QTG-seq, we fine-mapped a plant-height QTL in maize (Zea mays L.), qPH7, to a 300-kb genomic interval and verified that a gene encoding an NF-YC transcription factor was the functional gene. Functional analysis suggested that qPH7-encoding protein might influence plant height by interacting with a CO-like protein and an AP2 domain-containing protein. Selection footprint analysis indicated that qPH7 was subject to strong selection during maize improvement. In summary, QTG-seq provides an efficient method for QTL fine-mapping in the era of ‘‘big data’’.
Publication
Zhang HW, Wang X, Pan QC et al. QTG-seq accelerates QTL fine mapping through QTL partitioning and whole-genome sequencing of bulked segregant samples. Molecular Plant, 2018.